JupyterLab is an open source application built around the concept of a computational notebook document. It enables sharing and executing code, data processing, visualization, and offers a range of interactive features for creating graphs.
The latest version, JupyterLab 4.0, was released in early June. Compared to its predecessors, this version features a faster Web UI, improved editor performance, a new Extension Manager, and real-time collaboration.
If you have already installed the standalone 3.x version, evaluating the new features will require rewriting your current environment, which can be labor-intensive and risky. However, in environments where Docker operates, such as Docker Desktop, you can start an isolated JupyterLab 4.0 in a container without affecting your installed JupyterLab environment. Of course, you can run these without impacting the existing environment and access them on a different port.
In this article, we show how to quickly evaluate the new features of JupyterLab 4.0 using Jupyter Docker Stacks on Docker Desktop, without affecting the host PC side.

Why containerize JupyterLab?
Users have downloaded the base image of JupyterLab Notebook stack Docker Official Image more than 10 million times from Docker Hub. What’s driving this significant download rate? There’s an ever-increasing demand for Docker containers to streamline development workflows, while allowing JupyterLab developers to innovate with their choice of project-tailored tools, application stacks, and deployment environments. Our JupyterLab notebook stack official image also supports both AMD64 and Arm64/v8 platforms.
Containerizing the JupyterLab environment offers numerous benefits, including the following:
- Containerization ensures that your JupyterLab environment remains consistent across different deployments. Whether you’re running JupyterLab on your local machine, in a development environment, or in a production cluster, using the same container image guarantees a consistent setup. This approach helps eliminate compatibility issues and ensures that your notebooks behave the same way across different environments.
- Packaging JupyterLab in a container allows you to easily share your notebook environment with others, regardless of their operating system or setup. This eliminates the need for manually installing dependencies and configuring the environment, making it easier to collaborate and share reproducible research or workflows. And this is particularly helpful in AI/ML projects, where reproducibility is crucial.
- Containers enable scalability, allowing you to scale your JupyterLab environment based on the workload requirements. You can easily spin up multiple containers running JupyterLab instances, distribute the workload, and take advantage of container orchestration platforms like Kubernetes for efficient resource management. This becomes increasingly important in AI/ML development, where resource-intensive tasks are common.
Getting started
To use JupyterLab on your computer, one option is to use the JupyterLab Desktop application. It’s based on Electron, so it operates with a GUI on Windows, macOS, and Linux. Indeed, using JupyterLab Desktop makes the installation process fairly simple. In a Windows environment, however, you’ll also need to set up the Python language separately, and, to extend the capabilities, you’ll need to use pip to set up packages.
Although such a desktop solution may be simpler than building from scratch, we think the combination of Docker Desktop and Docker Stacks is still the more straightforward option. With JupyterLab Desktop, you cannot mix multiple versions or easily delete them after evaluation. Above all, it does not provide a consistent user experience across Windows, macOS, and Linux.
On a Windows command prompt, execute the following command to launch a basic notebook:
1 | docker container run -it --rm -p 10000:8888 jupyter/base-notebook |
This command utilizes the jupyter/base-notebook Docker image, maps the host’s port 10000 to the container’s port 8888, and enables command input and a pseudo-terminal. Additionally, an option is added to delete the container once the process is completed.
After waiting for the Docker image to download, access and token information will be displayed on the command prompt as follows. Here, rewrite the URL http://127.0.0.1:8888 to http://127.0.0.1:10000 and then append the token to the end of this URL. In this example, the output will look like this:
- Displayed on screen: http://127.0.0.1:8888/lab?token=6e302b2c99d56f1562e082091f4a3236051fb0a4135e10bb
- To be entered in the browser address: http://127.0.0.1:10000/lab?token=6e302b2c99d56f1562e082091f4a3236051fb0a4135e10bb
Note that this token is specific to my environment, so copying it will not work for you. You should replace it with the one actually displayed on your command prompt.
Then, after waiting for a short while, JupyterLab will launch (Figure 1). From here, you can start a Notebook, access Python’s console environment, or utilize other work environments.
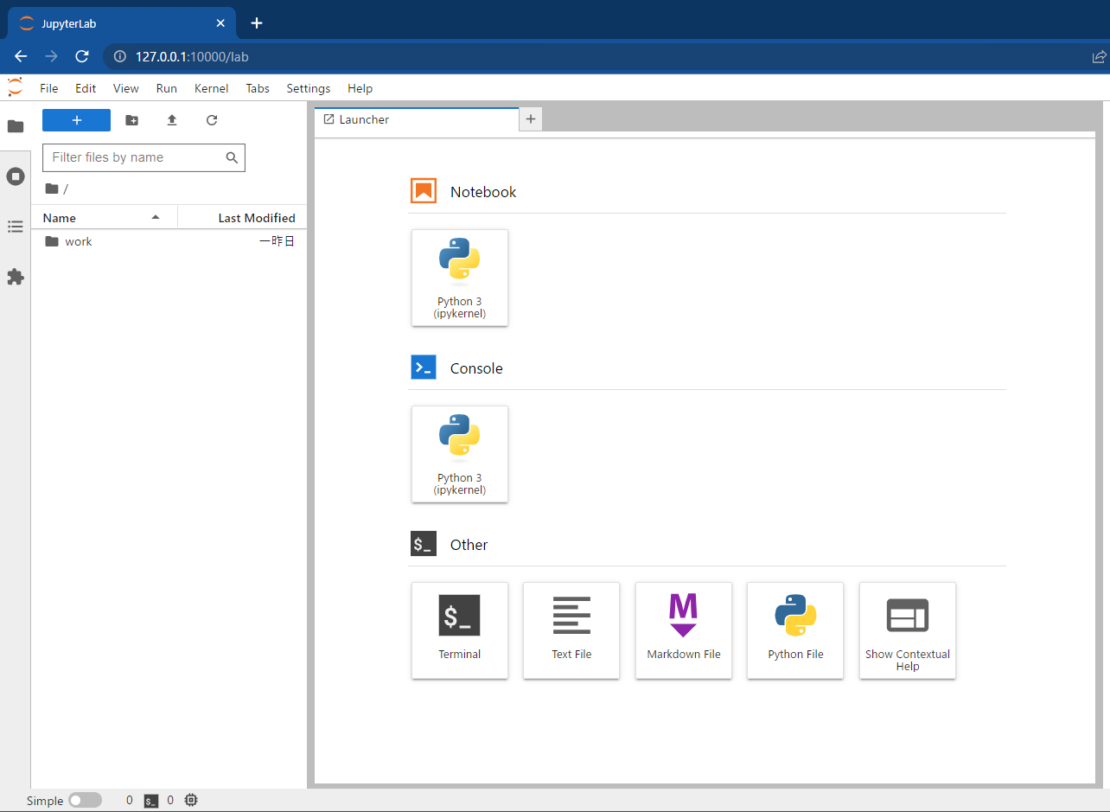
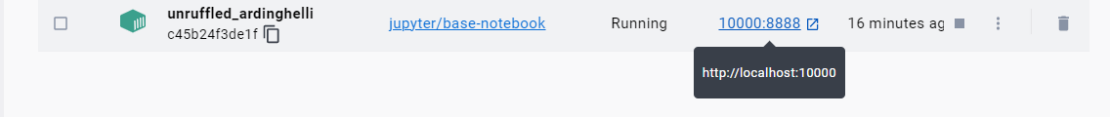
The port 10000 on the host side is mapped to port 8888 inside the container, as shown in Figure 2.
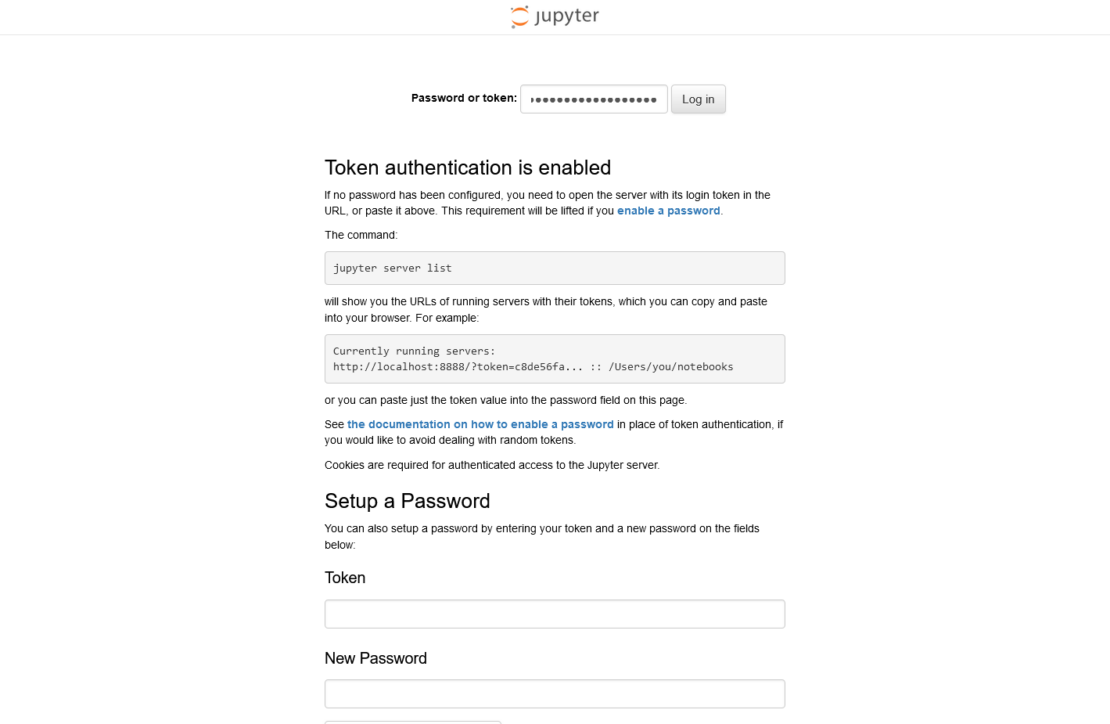
In the Password or token input form on the screen, enter the token displayed in the command line or in the container logs (the string following token=), and select Log in, as shown in Figure 3.
By the way, in this environment, the data will be erased when the container is stopped. If you want to reuse your data even after stopping the container, create a volume by adding the -v option when launching the Docker container.
To stop this container environment, click CTRL-C on the command prompt, then respond to the Jupyter server’s prompt Shutdown this Jupyter server (y/[n])? with y and press enter. If you are using Docker Desktop, stop the target container from the Containers.
1 2 3 4 5 | Shutdown this Jupyter server (y/[n])? y[C 2023-06-26 01:39:52.997 ServerApp] Shutdown confirmed[I 2023-06-26 01:39:52.998 ServerApp] Shutting down 5 extensions[I 2023-06-26 01:39:52.998 ServerApp] Shutting down 1 kernel[I 2023-06-26 01:39:52.998 ServerApp] Kernel shutdown: 653f7c27-03ff-4604-a06c-2cb4630c098d |
Once the display changes as follows, the container is terminated and the data is deleted.
When the container is running, data is saved in the /home/jovyan/work/ directory inside the container. You can either bind mount this as a volume or allocate it as a volume when starting the container. By doing so, even if you stop the container, you can use the same data again when you restart the container:
1 2 3 | docker container run -it -p 10000:8888 \ -v “%cd%”:/home/jovyan/work \ jupyter/base-notebook |
Note: The \ symbol signifies that the command line continues on the command prompt. You may also write the command in a single line without using the \ symbol. However, in the case of Windows command prompt, you need to use the ^ symbol instead.
With this setup, when launched, the JupyterLab container mounts the /work/ directory to the folder where the docker container run command was executed. Because the data persists even when the container is stopped, you can continue using your Notebook data as it is when you start the container again.
Plotting using the famous Iris flower dataset
In the following example, we’ll use the Iris flower dataset, which consists of 150 records in total, with 50 samples from each of three types of Iris flowers (Iris setosa, Iris virginica, Iris versicolor). Each record consists of four numerical attributes (sepal length, sepal width, petal length, petal width) and one categorical attribute (type of iris). This data is included in the Python library scikit-learn, and we will use matplotlib to plot this data.
When trying to input the sample code from the scikit-learn page (the code is at the bottom of the page, and you can copy and paste it) into iPython, the following error occurs (Figure 4).

This is an error message on iPython stating that the “matplotlib” module does not exist. Additionally, the “scikit-learn” module is needed.
To avoid these errors and enable plotting, run the following command. Here, !pip signifies running the pip command within the iPython environment:
1 | !pip install matplotlib scikit-learn |
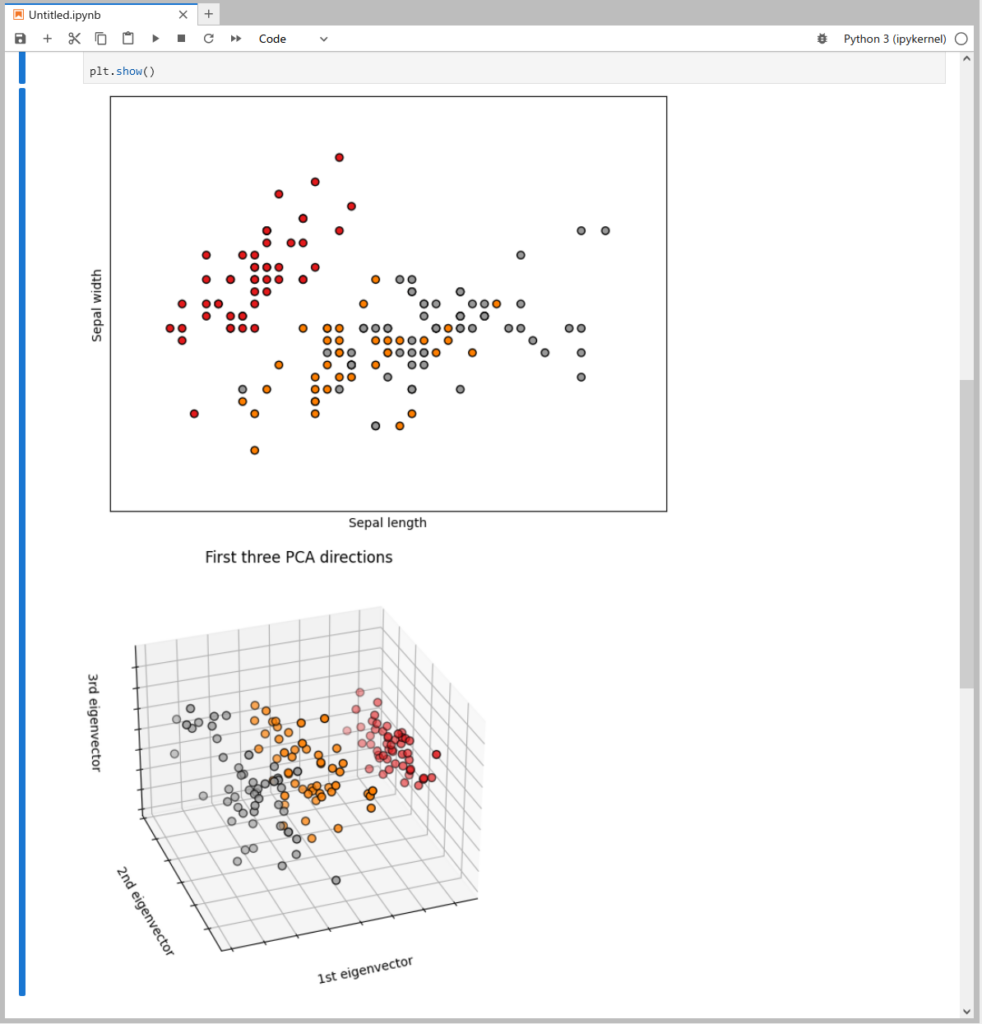
By pasting and executing the earlier sample code in the next cell on iPython, you can plot and display the Iris dataset as shown in Figure 5.
Note that it can be cumbersome to use the !pip command to add modules every time. Fortunately, you can add also add modules in the following ways:
- By creating a dedicated Dockerfile
- By using an existing group of images called Jupyter Docker Stacks
Building a Docker image
If you’re familiar with Dockerfile and building images, this five-step method is easy. Also, this approach can help keep the Docker image size in check.
Step 1. Creating a directory
To build a Docker image, the first step is to create and navigate to the directory where you’ll place your Dockerfile and context:
1 | mkdir myjupyter && cd myjupyter |
Step 2. Creating a requirements.txt file
Create a requirements.txt file and list the Python modules you want to add with the pip command:
1 2 | matplotlibscikit-learn |
Step 3. Writing a Dockerfile
1 2 3 | FROM jupyter/base-notebookCOPY ./requirements.txt /home/jovyan/workRUN python -m pip install --no-cache -r requirements.txt |
This Dockerfile specifies a base image jupyter/base-notebook, copies the requirements.txt file from the local directory to the /home/jovyan/work directory inside the container, and then runs a pip install command to install the Python packages listed in the requirements.txt file.
Step 4. Building the Docker image
1 | docker image build -t myjupyter |
Step 5. Launching the container
1 2 3 | docker container run -it -p 10000:8888 \ -v “%cd%”:/home/jovyan/work \ myjupyter |
Here’s what each part of this command does:
- The
docker runcommand instructs Docker to run a container. - The
-itoption attaches an interactive terminal to the container. - The
-p 10000:8888maps port 10000 on the host machine to port 8888 inside the container. This allows you to access Jupyter Notebook running in the container viahttp://localhost:10000in your web browser. - The
-v "%cd%":/home/jovyan/workmounts the current directory (%cd%) on the host machine to the/home/jovyan/workdirectory inside the container. This enables sharing files between the host and the Jupyter Notebook.
In this example, myjupyter is the name of the Docker image you want to run. Make sure you have the appropriate image available on your system. The operation after startup is the same as before. You don’t need to add libraries with the !pip command because the necessary libraries are included from the start.
How to use Jupyter Docker Stacks’ images
To execute the JupyterLab environment, we will utilize a Docker image called jupyter/scipy-notebook from the Jupyter Docker Stacks. Please note that the running Notebook will be terminated. After entering Ctrl-C on the command prompt, enter y and specify the running container.
Then, enter the following to run a new container:
1 2 3 | docker container run -it -p 10000:8888 \ -v “%cd%”:/home/jovyan/work \ jupyter/scipy-notebook |
This command will run a container using the jupyter/scipy-notebook image, which provides a Jupyter Notebook environment with additional scientific libraries.
Here’s a breakdown of the command:
- The
docker runcommand starts a new container. - The
-itoption attaches an interactive terminal to the container. - The
-p 10000:8888maps port 10000 on the host machine to port 8888 inside the container, allowing access to Jupyter Notebook at http://localhost:10000. - The
-v "$(pwd)":/home/jovyan/workmounts the current directory ($(pwd)) on the host machine to the/home/jovyan/workdirectory inside the container. This enables sharing files between the host and the Jupyter Notebook. - The
jupyter/scipy-notebookis the name of the Docker image used for the container. Make sure you have this image available on your system.
The previous JupyterLab image was a minimal Notebook environment. The image we are using this time includes many packages used in the scientific field, such as numpy and pandas, so it may take some time to download the Docker image. This one is close to 4GB in image size.
Once the container is running, you should be able to run the Iris dataset sample immediately without having to execute pip like before. Give it a try.
Some images include TensorFlow’s deep learning library, ones for the R language, Julia programming language, and Apache Spark. See the image list page for details.
In a Windows environment, you can easily run and evaluate the new version of JupyterLab 4.0 using Docker Desktop. Doing so will not affect or conflict with the existing Python language environment. Furthermore, this setup provides a consistent user experience across other platforms, such as macOS and Linux, making it the ideal solution for those who want to try it.
Conclusion
By containerizing JupyterLab with Docker, AI/ML developers gain numerous advantages, including consistency, easy sharing and collaboration, and scalability. It enables efficient management of AI/ML development workflows, making it easier to experiment, collaborate, and reproduce results across different environments. With JupyterLab 4.0 and Docker, the possibilities for supercharging your AI/ML development are limitless. So why wait? Embrace containerization and experience the true power of JupyterLab in your AI/ML projects.
References
Learn more
- Get the latest release of Docker Desktop.
- Vote on what’s next! Check out our public roadmap.
- Have questions? The Docker community is here to help.
- New to Docker? Get started.

